Introduction
PyTorch's DataLoader (torch.utils.data.Dataloader) is already a useful tool for efficiently loading and preprocessing data for training deep learning models. By default, PyTorch uses a single-worker process (num_workers=0), but users can specify a higher number to leverage parallelism and speed up data loading.
However, since it is a general-purpose dataloader, and even though it offers parallelization, it is still not suitable for certain custom use cases. In this post, we explore how we can speed up the loading of multiple 2D slices from a dataset of 3D medical scans using torch.multiprocessing().
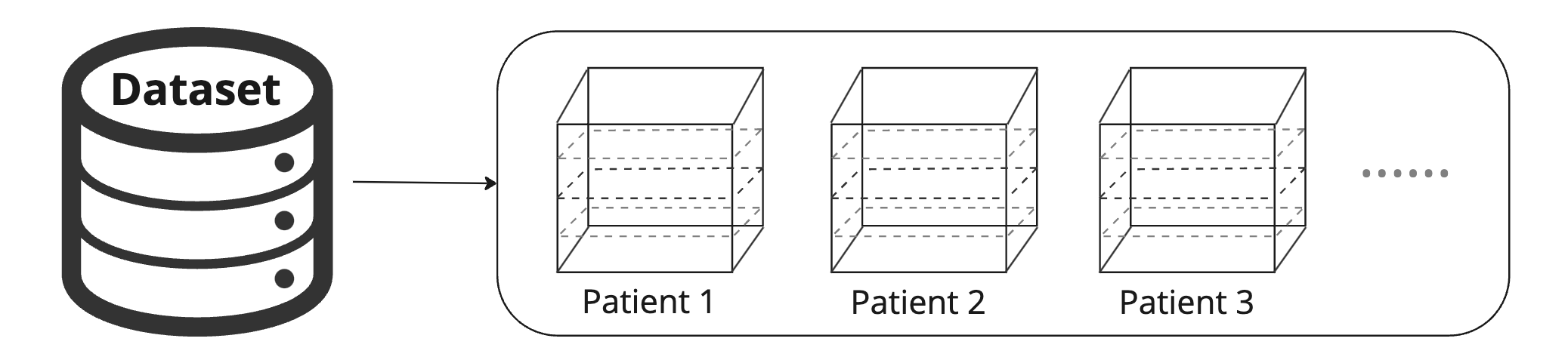
Our torch.utils.data.Dataset
Imagine a use case in which given a set of 3D scans for patients (i.e., P1, P2, P3, …) and a list of corresponding slices; our goal is to build a dataloader that outputs a slice in every iteration. Check the Python code below where we build a torch dataset called myDataset, and pass it into torch.utils.data.Dataloader().
# check full code here: https://gist.github.com/prerakmody/0c5e9263d42b2fab26a48dfb6b818cca#file-torchdataloader-py
import tqdm
import time
import torch # v1.12.1
import numpy as np
##################################################
# myDataset
##################################################
def getPatientArray(patientName):
# return patients 3D scan
def getPatientSliceArray(patientName, sliceId, patientArray=None):
# return patientArray and a slice
class myDataset(torch.utils.data.Dataset):
def __init__(self, patientSlicesList, patientsInMemory=1):
...
self.patientObj = {} # To store one patients 3D array. More patients lead to more memory usage.
def _managePatientObj(self, patientName):
if len(self.patientObj) > self.patientsInMemory:
self.patientObj.pop(list(self.patientObj.keys())[0])
def __getitem__(self, idx):
# Step 0 - Init
patientName, sliceId = ...
# Step 1 - Get patient slice array
patientArrayThis = self.patientObj.get(patientName, None)
patientArray, patientSliceArray = getPatientSliceArray(patientName, sliceId, patientArray=patientArrayThis)
if patientArray is not None:
self.patientObj[patientName] = patientArray
self._managePatientObj(patientName)
return patientSliceArray, [patientName, sliceId]
##################################################
# Main
##################################################
if __name__ == '__main__':
# Step 1 - Setup patient slices (fixed count of slices per patient)
patientSlicesList = {
'P1': [45, 62, 32, 21, 69]
, 'P2': [13, 23, 87, 54, 5]
, 'P3': [34, 56, 78, 90, 12]
, 'P4': [34, 56, 78, 90, 12]
}
workerCount, batchSize, epochs = 4, 1, 3
# Step 2.1 - Create dataset and dataloader
dataset = myDataset(patientSlicesList)
dataloader = torch.utils.data.DataLoader(dataset, batch_size=3, num_workers=4)
# Step 2.2 - Iterate over dataloader
print ('\n - [main] Iterating over (my) dataloader...')
for epochId in range(epochs):
print (' - [main] --------------------------------------- Epoch {}/{}'.format(epochId+1, epochs))
for i, (patientSliceArray, meta) in enumerate(dataloader):
print (' - [main] meta: ', meta)
pbar.update(patientSliceArray.shape[0])
The main concern with our use case is that 3D medical scans are large in size (emulated here by the time.sleep() operation) and hence
-
reading them from disk can be time intensive
-
and a large dataset of 3D scans in most cases cannot be pre-read into memory
Ideally, we should only read each patient scan once for all the slices associated with it. But since data is split by torch.utils.data.dataloader(myDataset, batch_size=b, workers=n) into workers depending on the batch size, there is a possibility for different workers to read a patient twice (check the image and log below).

- [main] Iterating over (my) dataloader...
- [main] --------------------------------------- Epoch 1/3
- [getPatientArray()][worker=3] Loading volumes for patient: P2
- [getPatientArray()][worker=1] Loading volumes for patient: P1
- [getPatientArray()][worker=2] Loading volumes for patient: P2
- [getPatientArray()][worker=0] Loading volumes for patient: P1
- [getPatientArray()][worker=3] Loading volumes for patient: P3
- [main] meta: [('P1', 'P1', 'P1'), tensor([45, 62, 32])]
- [getPatientArray()][worker=1] Loading volumes for patient: P2
- [main] meta: [('P1', 'P1', 'P2'), tensor([21, 69, 13])]
- [main] meta: [('P2', 'P2', 'P2'), tensor([23, 87, 54])]
- [main] meta: [('P2', 'P3', 'P3'), tensor([ 5, 34, 56])]
- [getPatientArray()][worker=2] Loading volumes for patient: P4
- [getPatientArray()][worker=0] Loading volumes for patient: P3
- [getPatientArray()][worker=1] Loading volumes for patient: P4
- [main] meta: [('P3', 'P3', 'P3'), tensor([78, 90, 12])]
- [main] meta: [('P4', 'P4', 'P4'), tensor([34, 56, 78])]
- [main] meta: [('P4', 'P4'), tensor([90, 12])]
To summarize, here are the issues with the existing implementation of torch.utils.data.Dataloader
- Each of the workers is passed a copy of the
myDataset()(Ref:torch v1.2.0 ), and since they do not have any shared memory, it leads to a double disk read of a patient’s 3D scan.
- Moreover, since the torch sequentially loops over
patientSliceList(see image below), no natural shuffling is possible between (patientId, sliceId) combos. (Note: one can shuffle, but that involves storing outputs in memory)

Note: One could also just return a bunch of slices together from each patients 3D scan. But if we wish to also return slice-dependent 3D arrays (for example, interactive refinement networks (see Fig1 of this work), then this greatly increases the memory footprint of your dataloader.
Using torch.multiprocessing
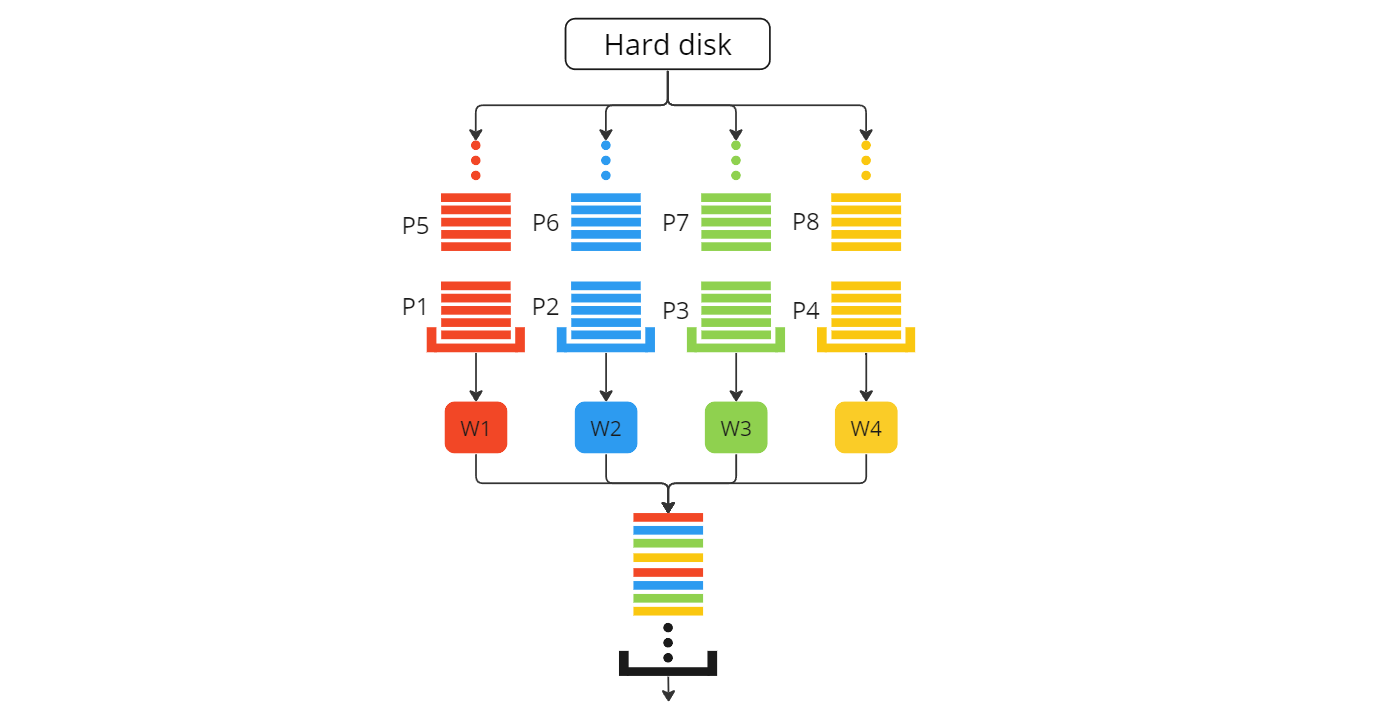
To prevent multiple reads of patient scans, we would ideally need each patient (let’s imagine 8 patients) to be read by a particular worker.

To achieve this, we use the same internal tools as the torch dataloader class (i.e., torch.multiprocessing()) but with a slight difference. Check the workflow figure and code below for our custom dataloader - myDataloader
# check full code here: https://gist.github.com/prerakmody/0c5e9263d42b2fab26a48dfb6b818cca#file-mydataloader-py
class myDataloader:
def __init__(self, patientSlicesList, numWorkers, batchSize) -> None:
...
self._initWorkers()
def _initWorkers(self):
# Step 1 - Initialize vas
self.workerProcesses = []
self.workerInputQueues = [torchMP.Queue() for _ in range(self.numWorkers)]
self.workerOutputQueue = torchMP.Queue()
for workerId in range(self.numWorkers):
p = torchMP.Process(target=getSlice, args=(workerId, self.workerInputQueues[workerId], self.workerOutputQueue))
p.start()
def fillInputQueues(self):
"""
This function allows to split patients and slices across workers. One can implement custom logic here.
"""
patientNames = list(self.patientSlicesList.keys())
for workerId in range(self.numWorkers):
idxs = ...
for patientName in patientNames[idxs]:
for sliceId in self.patientSlicesList[patientName]:
self.workerInputQueues[workerId].put((patientName, sliceId))
def emptyAllQueues(self):
# empties the self.workerInputQueues and self.workerOutputQueue
def __iter__(self):
try:
# Step 0 - Init
self.fillInputQueues() # once for each epoch
batchArray, batchMeta = [], []
# Step 1 - Continuously yield results
while True:
if not self.workerOutputQueue.empty():
# Step 2.1 - Get data point
patientSliceArray, patientName, sliceId = self.workerOutputQueue.get(timeout=QUEUE_TIMEOUT)
# Step 2.2 - Append to batch
...
# Step 2.3 - Yield batch
if len(batchArray) == self.batchSize:
batchArray = collate_tensor_fn(batchArray)
yield batchArray, batchMeta
batchArray, batchMeta = [], []
# Step 3 - End condition
if np.all([self.workerInputQueues[i].empty() for i in range(self.numWorkers)]) and self.workerOutputQueue.empty():
break
except GeneratorExit:
self.emptyAllQueues()
except KeyboardInterrupt:
self.closeProcesses()
except:
traceback.print_exc()
def closeProcesses(self):
pass
if __name__ == "__main__":
# Step 1 - Setup patient slices (fixed count of slices per patient)
patientSlicesList = {
'P1': [45, 62, 32, 21, 69]
, 'P2': [13, 23, 87, 54, 5]
, 'P3': [34, 56, 78, 90, 12]
, 'P4': [34, 56, 78, 90, 12]
, 'P5': [45, 62, 32, 21, 69]
, 'P6': [13, 23, 87, 54, 5]
, 'P7': [34, 56, 78, 90, 12]
, 'P8': [34, 56, 78, 90, 12, 21]
}
workerCount, batchSize, epochs = 4, 1, 3
# Step 2 - Create new dataloader
dataloaderNew = None
try:
dataloaderNew = myDataloader(patientSlicesList, numWorkers=workerCount, batchSize=batchSize)
print ('\n - [main] Iterating over (my) dataloader...')
for epochId in range(epochs):
with tqdm.tqdm(total=len(dataset), desc=' - Epoch {}/{}'.format(epochId+1, epochs)) as pbar:
for i, (X, meta) in enumerate(dataloaderNew):
print (' - [main] {}'.format(meta.tolist()))
pbar.update(X.shape[0])
dataloaderNew.closeProcesses()
except KeyboardInterrupt:
if dataloader is not None: dataloader.closeProcesses()
except:
traceback.print_exc()
if dataloaderNew is not None: dataloaderNew.closeProcesses()
The snippet above (with 8 patients instead) contains the following functions
__iter__()- SincemyDataloader()is a loop, this is the function it actually loops over.
_initWorkers()- Here, we create our worker processes with their individual input queuesworkerInputQueues[workerId]. This is called when the class is initialized.
fillInputQueues()- This function is called when we begin the loop (essentially at the start of every epoch). It fills up the individual worker’s input queue.
getSlice()- This is the main logic function that returns a slice from a patient volume. Check the code here.
collate_tensor_fn()- This function is directly copied from the torch repo - torchv1.12.0 and is used to batch data together.
Performance
To test whether our dataloader offers a speedup compared to the default option, we test the speed of each dataloader loop using different worker counts. We varied two parameters in our experiments:
Number of Workers : We tested 1, 2, 4, and 8 worker processes.Batch Size : We evaluated different batch sizes ranging from 1 to 8.
Toy Dataset
We first experiment with our toy dataset and see that our dataloader performs much faster. See the figure below (or reproduce with this code)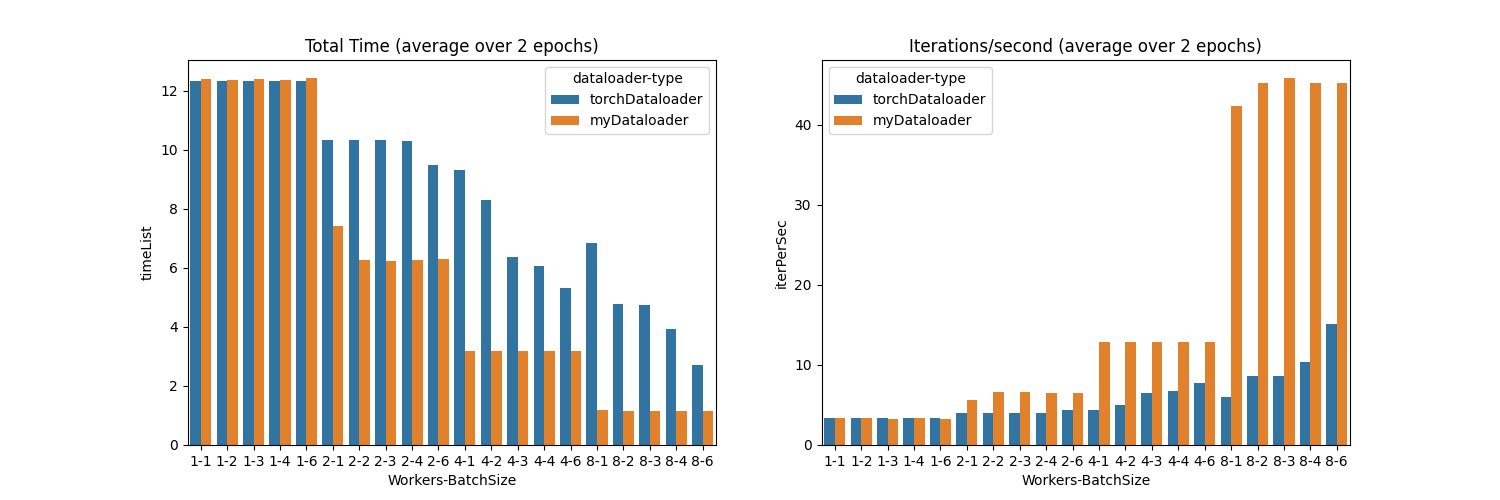
Here, we can see the following
- When using a single worker, both dataloaders are the same.
- When using additional workers (i.e. 2,4,8), there is a speedup in both dataloaders, however, the speedup is much higher in our custom dataloader.
- When using a batch size of 6 (as compared to 1,2,3,4), there is a small hit in the performance. This is because, in our toy dataset, the
patientSlicesListvariable contains 5 slices per patient. So, the worker needs to wait to read the second patient to add to the last index of the batch.
Real World Dataset
We then benchmark a real dataset where 3D scans are loaded, a slice is extracted,
We observed that
Resource Utilization
We also monitored resource utilization during data loading with varying worker counts. With a higher number of workers, we observed increased CPU and memory usage, which is expected due to the parallelism introduced by additional processes. Users should consider their hardware constraints and resource availability when choosing the optimal worker count.
Summary
-
In this blog post, we explored the limitations of PyTorch's standard DataLoader when dealing with datasets containing large 3D medical scans and presented a custom solution using
torch.multiprocessingto improve data loading efficiency. -
In the context of slice extraction from these 3D medical scans, the default dataLoader can potentially lead to multiple reads of the same patient scan as workers do not share memory. This redundancy causes significant delays, particularly when dealing with large datasets.
-
Our custom dataLoader splits patients between workers, ensuring that each 3D scan is read only once per worker. This approach prevents redundant disk reads and leverages parallel processing to speed up data loading.
-
Performance testing showed that our custom dataLoader generally outperforms the standard dataLoader, especially with smaller batch sizes and multiple worker processes.
- However, the performance gains diminished with larger batch sizes.
Our custom dataLoader enhances data loading efficiency for large 3D medical datasets by reducing redundant reads and maximizing parallelism. This improvement can lead to faster training times and better utilization of hardware resources.
This blog was written together with my colleague Jingnan Jia.